Download Web Page In R
The Department of Criminal Justice in Texas keeps records of every inmate they execute. This tutorial will show you how to scrape that data, which lives in a table on the website and download the images.
The tutorial uses rvest and xml to scrape tables, purrr to download and export files, and magick to manipulate images. For an introduction to R Studio go here and for help with dplyr go here.
Scraping the data from HTML websites
Load the xml2 package and define the url with the data (here it's webpage_url).
library(xml2) webpage_url <- "http://www.tdcj.state.tx.us/death_row/dr_executed_offenders.html" webpage <- xml2 ::read_html(webpage_url)
Use the rvest::html_table() and tibble::as_tibble() to put the content into a tibble.
ExOffndrsRaw <- rvest ::html_table(webpage)[[1]] %>% tibble ::as_tibble(.name_repair = "unique" ) # repair the repeated columns ExOffndrsRaw %>% dplyr ::glimpse(45)
Observations: 555 Variables: 10 $ Execution <int> 555, 554, 553, 552, 5… $ Link..2 <chr> "Offender Information… $ Link..3 <chr> "Last Statement", "La… $ `Last Name` <chr> "Acker", "Clark", "Yo… $ `First Name` <chr> "Daniel", "Troy", "Ch… $ TDCJNumber <int> 999381, 999351, 99950… $ Age <int> 46, 51, 34, 66, 37, 3… $ Date <chr> "9/27/2018", "9/26/20… $ Race <chr> "White", "White", "Bl… $ County <chr> "Hopkins", "Smith", "… Assign some new names and do a little wrangling
The Link..2 and Link..3 variables don't contain the actual links,
ExOffndrsRaw %>% dplyr ::select(dplyr ::contains( 'Link' )) %>% head(5)
# A tibble: 5 x 2 Link..2 Link..3 <chr> <chr> 1 Offender Information Last Statement 2 Offender Information Last Statement 3 Offender Information Last Statement 4 Offender Information Last Statement 5 Offender Information Last Statement …so we will have to go back to the webpage to find these. It also looks like the Race variable has a misspelling.
ExOffndrsRaw %>% dplyr ::count(Race)
# A tibble: 5 x 2 Race n <chr> <int> 1 Black 201 2 Hispanic 104 3 Histpanic 1 4 Other 2 5 White 247 Collapse the Histpanic level into Hispanic.
# new var names ExOffndrs_names <- c( "execution" , "url_offndr" , "url_lststmnt" , "last_name" , "first_name" , "tdcj_number" , "age" , "date" , "race" , "county" ) # assign new names ExOffndrs <- ExOffndrsRaw %>% magrittr ::set_names(ExOffndrs_names) %>% # reorganize a bit dplyr ::select(first_name, last_name, race, age, date, county, execution, url_offndr, url_lststmnt) %>% # format date dplyr ::mutate(date = lubridate ::mdy(date), # fix up the race variable race = dplyr ::case_when( race == "Histpanic" ~ "Hispanic" , race == "Black" ~ "Black" , race == "Hispanic" ~ "Hispanic" , race == "White" ~ "White" , race == "Other" ~ "Other" )) # remove links ExOffndrs <- ExOffndrs %>% dplyr ::select(- dplyr ::contains( 'Link' )) ExOffndrs %>% dplyr ::glimpse(45)
Observations: 555 Variables: 9 $ first_name <chr> "Daniel", "Troy", "Ch… $ last_name <chr> "Acker", "Clark", "Yo… $ race <chr> "White", "White", "Bl… $ age <int> 46, 51, 34, 66, 37, 3… $ date <date> 2018-09-27, 2018-09-… $ county <chr> "Hopkins", "Smith", "… $ execution <int> 555, 554, 553, 552, 5… $ url_offndr <chr> "Offender Information… $ url_lststmnt <chr> "Last Statement", "La… Identify the links to offender information and last statements
Identify the links using the selector gadget.
Download the selector gadget app for your browser.
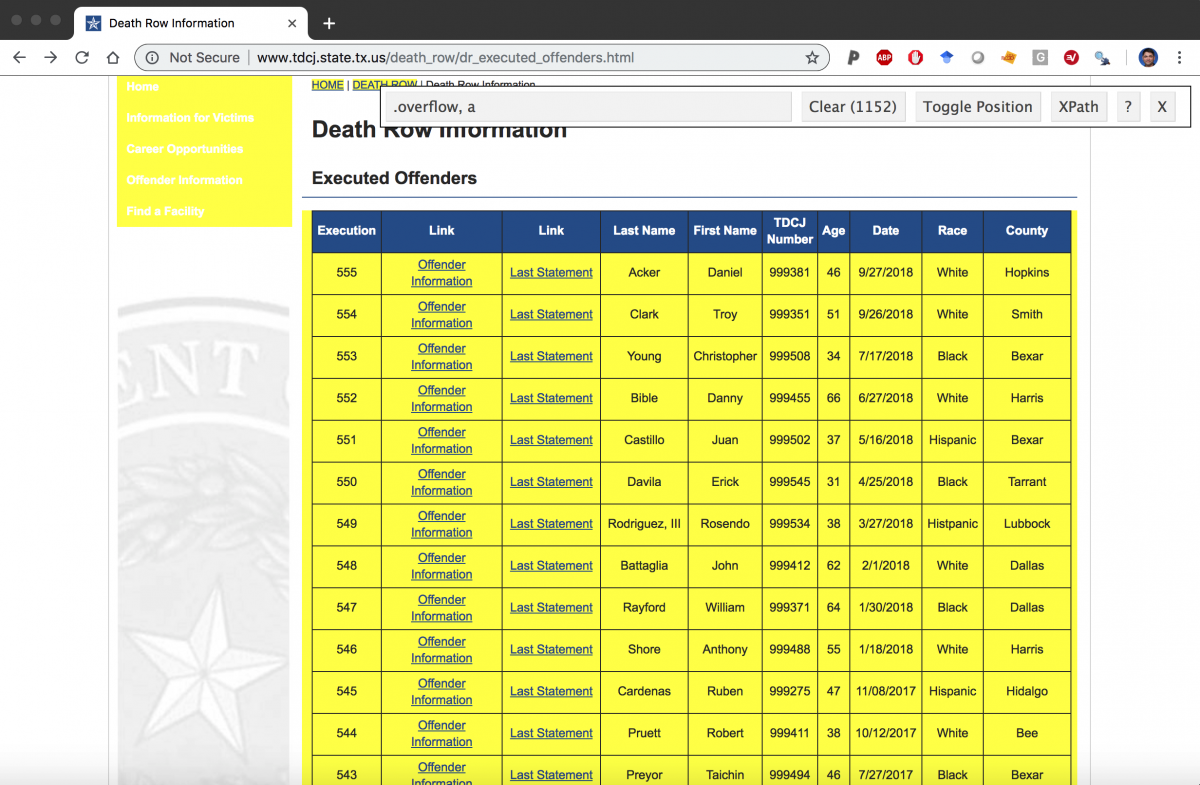
In order to get the links from the table, we need to pass webpage through a few passes of rvest functions (html_nodes and html_attr) with various css tags to get the correct url paths. This takes some trial and error, but eventually I was able to figure out the the correct combinations to get the links to the pages.
Links <- webpage %>% # this get the links in the overflow table # row rvest ::html_nodes( ".overflow tr" ) %>% # the links rvest ::html_nodes( "a" ) %>% # the header ref rvest ::html_attr( "href" ) # check Links Links %>% utils ::head(20)
[1] "dr_info/ackerdaniel.html" [2] "dr_info/ackerdaniellast.html" [3] "dr_info/clarktroy.html" [4] "dr_info/clarktroylast.html" [5] "dr_info/youngchristopher.html" [6] "dr_info/youngchristopherlast.html" [7] "dr_info/bibledanny.html" [8] "dr_info/bibledannylast.html" [9] "dr_info/castillojuan.html" [10] "dr_info/castillojuanlast.html" [11] "dr_info/davilaerick.html" [12] "dr_info/davilaericklast.html" [13] "dr_info/rodriguezrosendo.html" [14] "dr_info/rodriguezrosendolast.html" [15] "dr_info/battagliajohn.html" [16] "dr_info/battagliajohnlast.html" [17] "dr_info/rayfordwilliam.html" [18] "dr_info/rayfordwilliamlast.html" [19] "dr_info/shoreanthony.html" [20] "dr_info/shoreanthonylast.html" The Links contain:
- A
dr_info/path (which makes the entire path"http://www.tdcj.state.tx.us/death_row/dr_info/"). - Every offender has two links–one with their full name, the other with a
laststring attached to the back of their full name.
Something tells me if I check the base::length() of Links with the base::nrow()s in ExOffndrs…there will be twice as many links as rows in executed offenders.
[1] 1110 [1] 555 Good–this is what I want. That means each row in ExOffndrs has two links associated with their name.
Clean up the last statements
The stringr package can help me wrangle this long vector into the ExOffndrs tibble.
last_pattern <- stringr ::str_detect( string = Links, pattern = "last" ) utils ::head(Links[last_pattern])
[1] "dr_info/ackerdaniellast.html" "dr_info/clarktroylast.html" [3] "dr_info/youngchristopherlast.html" "dr_info/bibledannylast.html" [5] "dr_info/castillojuanlast.html" "dr_info/davilaericklast.html" Check to see that Links[last_pattern] is same length as the number of rows in ExOffndrs…
base ::identical(x = base ::length( Links[last_pattern]), y = base ::nrow( ExOffndrs))
[1] TRUE Subset the Links for the last_pattern, then tidy this vector into a tibble column. Finally, add the first portion of the url used above.
last_links <- Links[last_pattern] last_links <- last_links %>% tibble ::as_tibble(.name_repair = "unique" ) %>% tidyr ::gather(key = "key" , value = "value" ) %>% dplyr ::select(- key, last_url = value) %>% dplyr ::mutate( last_url = paste0( "http://www.tdcj.state.tx.us/death_row/" , last_url)) last_links %>% utils ::head(10)
# A tibble: 10 x 1 last_url <chr> 1 http://www.tdcj.state.tx.us/death_row/dr_info/ackerdaniellast.html 2 http://www.tdcj.state.tx.us/death_row/dr_info/clarktroylast.html 3 http://www.tdcj.state.tx.us/death_row/dr_info/youngchristopherlast.html 4 http://www.tdcj.state.tx.us/death_row/dr_info/bibledannylast.html 5 http://www.tdcj.state.tx.us/death_row/dr_info/castillojuanlast.html 6 http://www.tdcj.state.tx.us/death_row/dr_info/davilaericklast.html 7 http://www.tdcj.state.tx.us/death_row/dr_info/rodriguezrosendolast.html 8 http://www.tdcj.state.tx.us/death_row/dr_info/battagliajohnlast.html 9 http://www.tdcj.state.tx.us/death_row/dr_info/rayfordwilliamlast.html 10 http://www.tdcj.state.tx.us/death_row/dr_info/shoreanthonylast.html last_links %>% utils ::tail(10)
# A tibble: 10 x 1 last_url <chr> 1 http://www.tdcj.state.tx.us/death_row/dr_info/rumbaughcharleslast.html 2 http://www.tdcj.state.tx.us/death_row/dr_info/porterhenrylast.html 3 http://www.tdcj.state.tx.us/death_row/dr_info/miltoncharleslast.html 4 http://www.tdcj.state.tx.us/death_row/dr_info/no_last_statement.html 5 http://www.tdcj.state.tx.us/death_row/dr_info/morinstephenlast.html 6 http://www.tdcj.state.tx.us/death_row/dr_info/skillerndoylelast.html 7 http://www.tdcj.state.tx.us/death_row/dr_info/barefootthomaslast.html 8 http://www.tdcj.state.tx.us/death_row/dr_info/obryanronaldlast.html 9 http://www.tdcj.state.tx.us/death_row/dr_info/no_last_statement.html 10 http://www.tdcj.state.tx.us/death_row/dr_info/brookscharlielast.html Test one of these out in the browser.
Clean up the dr_info
Now I want the offender information links (so I omit the links with last in the pattern).
info_pattern <- ! stringr ::str_detect( string = Links, pattern = "last" ) utils ::head(Links[info_pattern])
[1] "dr_info/ackerdaniel.html" "dr_info/clarktroy.html" [3] "dr_info/youngchristopher.html" "dr_info/bibledanny.html" [5] "dr_info/castillojuan.html" "dr_info/davilaerick.html" Check the base::length().
base ::identical(x = base ::length( Links[info_pattern]), y = base ::nrow( ExOffndrs))
[1] TRUE Repeat the process from above.
info_links <- Links[info_pattern] info_links <- info_links %>% tibble ::as_tibble( .name_repair = "unique" ) %>% tidyr ::gather(key = "key" , value = "value" ) %>% dplyr ::select(- key, info_url = value) %>% dplyr ::mutate( info_url = paste0( "http://www.tdcj.state.tx.us/death_row/" , info_url)) info_links %>% utils ::head(10)
# A tibble: 10 x 1 info_url <chr> 1 http://www.tdcj.state.tx.us/death_row/dr_info/ackerdaniel.html 2 http://www.tdcj.state.tx.us/death_row/dr_info/clarktroy.html 3 http://www.tdcj.state.tx.us/death_row/dr_info/youngchristopher.html 4 http://www.tdcj.state.tx.us/death_row/dr_info/bibledanny.html 5 http://www.tdcj.state.tx.us/death_row/dr_info/castillojuan.html 6 http://www.tdcj.state.tx.us/death_row/dr_info/davilaerick.html 7 http://www.tdcj.state.tx.us/death_row/dr_info/rodriguezrosendo.html 8 http://www.tdcj.state.tx.us/death_row/dr_info/battagliajohn.html 9 http://www.tdcj.state.tx.us/death_row/dr_info/rayfordwilliam.html 10 http://www.tdcj.state.tx.us/death_row/dr_info/shoreanthony.html info_links %>% utils ::tail(10)
# A tibble: 10 x 1 info_url <chr> 1 http://www.tdcj.state.tx.us/death_row/dr_info/rumbaugh.jpg 2 http://www.tdcj.state.tx.us/death_row/dr_info/porterhenry.jpg 3 http://www.tdcj.state.tx.us/death_row/dr_info/miltoncharles.jpg 4 http://www.tdcj.state.tx.us/death_row/dr_info/delarosajesse.jpg 5 http://www.tdcj.state.tx.us/death_row/dr_info/morinstephen.jpg 6 http://www.tdcj.state.tx.us/death_row/dr_info/skillerndoyle.jpg 7 http://www.tdcj.state.tx.us/death_row/dr_info/barefootthomas.jpg 8 http://www.tdcj.state.tx.us/death_row/dr_info/obryanronald.jpg 9 http://www.tdcj.state.tx.us/death_row/dr_info/autryjames.html 10 http://www.tdcj.state.tx.us/death_row/dr_info/brookscharlie.html Note the different file extensions!
Some of the urls end in .html, others are .jpgs. Test a few of these out in the browser:
This is an .html file.
http://www.tdcj.state.tx.us/death_row/dr_info/brookscharlie.html
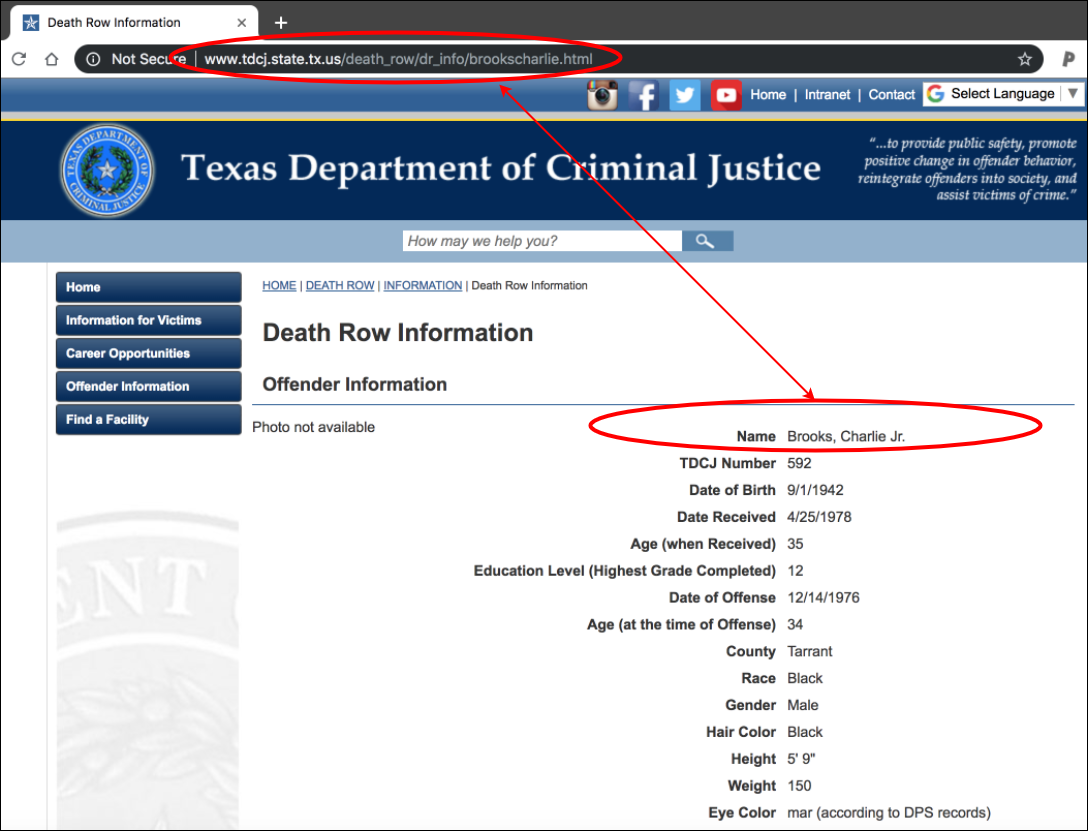
Now we assign these links to the ExOffndrs data frame. But first make sure they match up.
ExOffndrs %>% dplyr ::select(last_name, first_name) %>% utils ::head(10)
# A tibble: 10 x 2 last_name first_name <chr> <chr> 1 Acker Daniel 2 Clark Troy 3 Young Christopher 4 Bible Danny 5 Castillo Juan 6 Davila Erick 7 Rodriguez, III Rosendo 8 Battaglia John 9 Rayford William 10 Shore Anthony ExOffndrs %>% dplyr ::select(last_name, first_name) %>% utils ::tail(10)
# A tibble: 10 x 2 last_name first_name <chr> <chr> 1 Rumbaugh Charles 2 Porter Henry 3 Milton Charles 4 De La Rosa Jesse 5 Morin Stephen 6 Skillern Doyle 7 Barefoot Thomas 8 O'Bryan Ronald 9 Autry James 10 Brooks, Jr. Charlie Use dplyr::bind_cols() to attach these columns to ExOffndrs and rename to ExOffndrsComplete
ExOffndrsComplete <- ExOffndrs %>% dplyr ::bind_cols(last_links) %>% dplyr ::bind_cols(info_links) %>% dplyr ::select(dplyr ::ends_with( "name" ), last_url, info_url, dplyr ::everything(), - url_offndr, - url_lststmnt) ExOffndrsComplete %>% dplyr ::glimpse(78)
Observations: 555 Variables: 9 $ first_name <chr> "Daniel", "Troy", "Christopher", "Danny", "Juan", "Erick… $ last_name <chr> "Acker", "Clark", "Young", "Bible", "Castillo", "Davila"… $ last_url <chr> "http://www.tdcj.state.tx.us/death_row/dr_info/ackerdani… $ info_url <chr> "http://www.tdcj.state.tx.us/death_row/dr_info/ackerdani… $ race <chr> "White", "White", "Black", "White", "Hispanic", "Black",… $ age <int> 46, 51, 34, 66, 37, 31, 38, 62, 64, 55, 47, 38, 46, 61, … $ date <date> 2018-09-27, 2018-09-26, 2018-07-17, 2018-06-27, 2018-05… $ county <chr> "Hopkins", "Smith", "Bexar", "Harris", "Bexar", "Tarrant… $ execution <int> 555, 554, 553, 552, 551, 550, 549, 548, 547, 546, 545, 5… Create indicator for .html vs .jpgs
Create a binary variable to identify if this is a.jpg or.html path and name the new data frameExOffndrsComplete.
ExOffndrsComplete <- ExOffndrs %>% dplyr ::mutate(jpg_html = dplyr ::case_when( str_detect(string = info_url, pattern = ".jpg" ) ~ "jpg" , str_detect(string = info_url, pattern = ".html" ) ~ "html" ))
Usedplyr::sample_n to check a few examples of this new variable.
ExOffndrsComplete %>% dplyr ::sample_n(size = 10) %>% dplyr ::select(info_url, jpg_html) %>% utils ::head(10)
# A tibble: 10 x 2 info_url jpg_html <chr> <chr> 1 http://www.tdcj.state.tx.us/death_row/dr_info/richardsonjames… jpg 2 http://www.tdcj.state.tx.us/death_row/dr_info/canturuben.html html 3 http://www.tdcj.state.tx.us/death_row/dr_info/vuonghai.jpg jpg 4 http://www.tdcj.state.tx.us/death_row/dr_info/ramirezluis.html html 5 http://www.tdcj.state.tx.us/death_row/dr_info/beetsbetty.jpg jpg 6 http://www.tdcj.state.tx.us/death_row/dr_info/reeselamont.html html 7 http://www.tdcj.state.tx.us/death_row/dr_info/landryraymond.j… jpg 8 http://www.tdcj.state.tx.us/death_row/dr_info/tuttlecharles.j… jpg 9 http://www.tdcj.state.tx.us/death_row/dr_info/lawtonstacey.jpg jpg 10 http://www.tdcj.state.tx.us/death_row/dr_info/mannsdenard.html html Check count to be extra sure.
ExOffndrsComplete %>% dplyr ::count(jpg_html)
# A tibble: 2 x 2 jpg_html n <chr> <int> 1 html 177 2 jpg 378 Use purrr to download the .jpg files
Follow these three purrr steps from the workshop by Charlotte Wickham.
1. Do it for one element
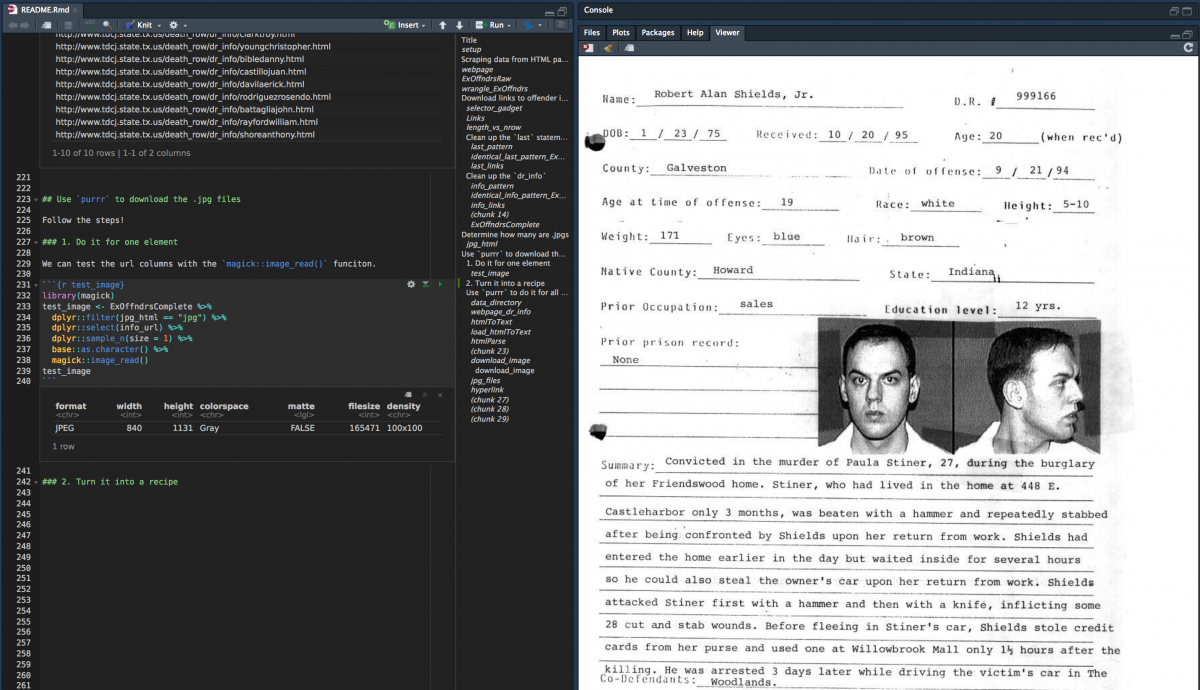
We can test the new url columns in the ExOffndrsComplete with the magick::image_read() funciton.
library(magick) test_image <- ExOffndrsComplete %>% dplyr ::filter(jpg_html == "jpg" ) %>% dplyr ::select(info_url) %>% dplyr ::sample_n(size = 1) %>% base ::as.character() magick ::image_read(test_image)
You should see an image in the RStudio viewer pane.
2. Turn it into a recipe
Put these urls into a vector (jpg_url), then create a folder to download them into (jpg_path).
ExOffndrsCompleteJpgs <- ExOffndrsComplete %>% dplyr ::filter(jpg_html == "jpg" ) jpg_url <- ExOffndrsCompleteJpgs $ info_url if (! base ::file.exists( "jpgs/" )) { base ::dir.create( "jpgs/" ) } jpg_path <- paste0( "jpgs/" , base ::basename(jpg_url))
3. Use purrr to download all files
Now use the purrr::walk2() function to download the files. How does walk2 work?
First look at the arguments for utils::download.file().
base ::args(utils :: download.file)
function (url, destfile, method, quiet = FALSE, mode = "w", cacheOK = TRUE, extra = getOption("download.file.extra"), ...) NULL How to walk2()
The help files tell us this function is "specialised for the two argument case".
So .x and .y become the two arguments we need to iterate over download.file(),
-
.x= the file path, which we created with the selector gadget above (injpg_url) -
.y= the location we want the files to end up (jpg_path), and - the function we want to iterate over
.xand.y(download.file).
This tells R to go the url, download the file located there, and put it in the associated /jpgs folder.
Download .jpg files
purrr ::walk2(.x = jpg_url, .y = jpg_path, .f = download.file) # trying URL 'http://www.tdcj.state.tx.us/death_row/dr_info/bigbyjames.jpg' # Content type 'image/jpeg' length 129128 bytes (126 KB) # ================================================== # downloaded 126 KB # # trying URL 'http://www.tdcj.state.tx.us/death_row/dr_info/ruizroland.jpg' # Content type 'image/jpeg' length 138981 bytes (135 KB) # ================================================== # downloaded 135 KB # # trying URL 'http://www.tdcj.state.tx.us/death_row/dr_info/garciagustavo.jpg' # Content type 'image/jpeg' length 164678 bytes (160 KB) # ================================================== # downloaded 160 KB
This might take awhile, but when its done, check the number of files in this folder.
fs ::dir_ls( "jpgs" ) %>% base ::length()
There you have it! 377 images of downloaded offenders!
Usepurrr anddplyr to split and export .csv files
This next use ofpurrr and iteration will cover how to:
- Split the
ExOffndrsCompletedata frame intoExExOffndrshtmlandExExOffndrsjpg - Save each of these data frames as .csv files
We should have two datasets with the following counts.
ExOffndrsComplete %>% dplyr ::count(jpg_html, sort = TRUE)
# A tibble: 2 x 2 jpg_html n <chr> <int> 1 jpg 378 2 html 180 These are new experimental functions fromdplyr, and a big shout out to Luis Verde Arregoitia for his post on a similar topic.
Thedplyr::group_split()"returns a list of tibbles. Each tibble contains the rows of .tbl for the associated group and all the columns, including the grouping variables", and I combine it withpurrr::walk() andreadr::write_csv() to export each file.
# fs::dir_ls(".") if (!file.exists( "data" )) { dir.create( "data" ) } ExOffndrsComplete %>% dplyr ::group_by(jpg_html) %>% dplyr ::group_split() %>% purrr ::walk(~.x %>% write_csv(path = paste0( "data/" , base ::noquote(lubridate ::today()), "-ExExOffndrs" , unique(.x $ jpg_html), ".csv" ))) fs ::dir_ls( "data" )
data/2018-12-20-ExExOffndrshtml.csv data/2018-12-20-ExExOffndrsjpg.csv data/2018-12-20-ExOffndrsComplete.csv - Author
- Recent Posts

Source: https://www.storybench.org/scraping-html-tables-and-downloading-files-with-r/
Posted by: goeswith.blogspot.com